Research Focus:
Develop and apply high-throughput single cell and spatial multiomics sequencing technologies to investigate tumor origin and evolution, and AI to model cell dynamics.
Research Work:
Understanding the origin and evolution of tumor cells is crucial for comprehending cancer initiation, recurrence, and for guiding subsequent tumor prevention and treatment strategies. Tumor cells arise from the accumulation of mutations within normal cells and then undergo multiple stages of evolution under various selective pressures, including the microenvironment, surgery, radiotherapy, chemotherapy, and immunotherapy. Studying this evolutionary process can unveil the mechanisms of cancer occurrence and progression, identify genome-related events tied to the origin and evolution of cancer cells, and ultimately provide new therapeutic targets.
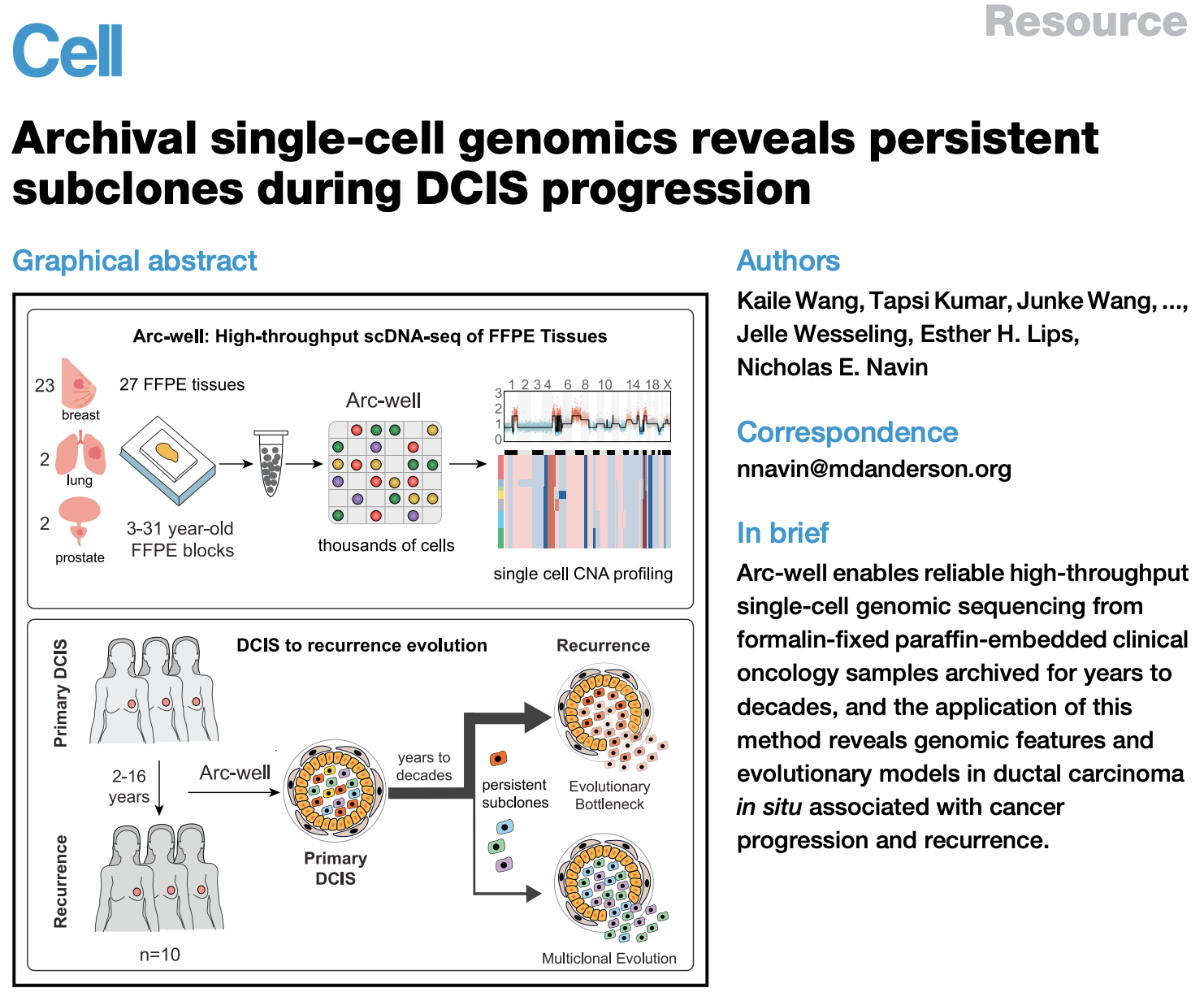
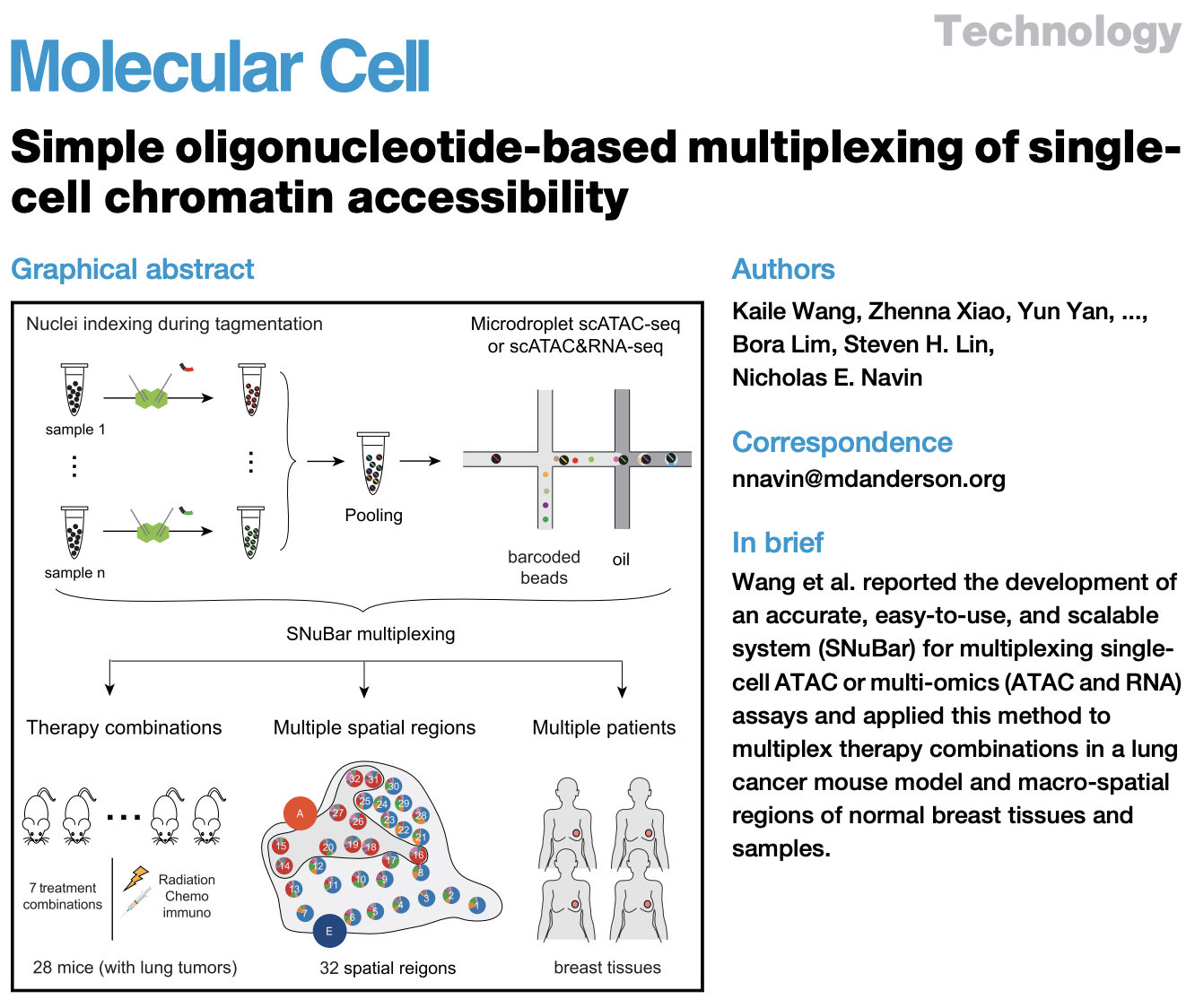
In previous work, our research group developed Arc-well, the first high-throughput single cell DNA sequencing technology that is compatible with FFPE tissues; SNuBar, a method for labeling the spatial location or sample information of single cells, as well as multiple high-throughput sequencing-based low-frequency mutation detection methods (o2n-seq, Droplet-CirSeq, Oseq, and EasyMF) to systematically study tumor evolution. These studies identified persistent subclones during the progression of primary ductal carcinoma in situ (DCIS) to breast cancer recurrence, revealed a dominant evolutionary bottleneck model, and identified genomic variations associated with breast cancer recurrence, providing new therapeutic targets.
Our group focuses on developing and applying high-throughput single cell multi-omics and spatial sequencing technologies to address fundamental questions on tumor cell origin and evolution, including the epithelial cell-of-origin of tumor cells, the role of somatic mutations in cancer initiation, the spatial localization of tumor subclones, their interactions with the microenvironment, and the relationship between aging and cancer. Beyond cancer, we are fascinated by using the unique single-cell multi-omics datasets we are producing to digitize single cells, further exploring the underlying principles of cell behaviors.